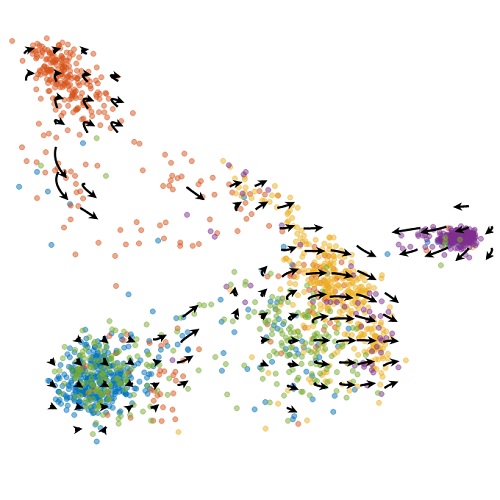Statistics
Leveraging multiomics to discover biology
In brief: I develop methods and analyses for multiomic data, with a particular focus on understanding its benefits of standard scRNA-seq.
Over the past decade, the increasing affordability of DNA sequencing has made RNA quantification more practical and spurred improvements in sequencing. Yet transcriptomics is only part of the sought-after comprehensive picture of single-cell biology. To hone understanding of biological regulation, it is necessary to quantify more modalities involved in information transfer: DNA and protein molecules, as well as transient and alternative RNA isoforms.
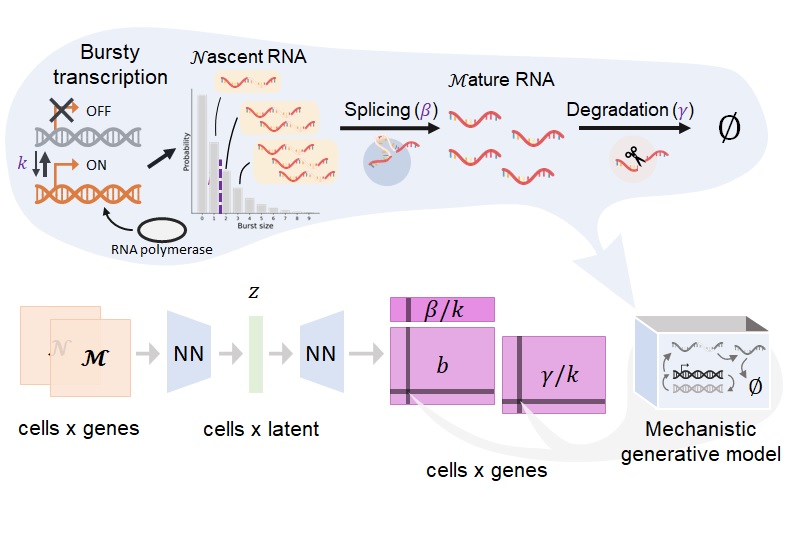
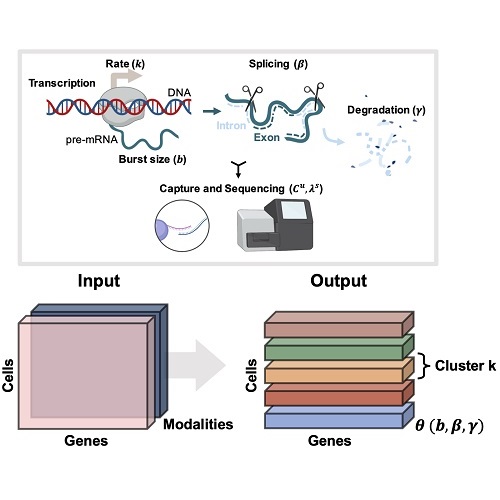
Simultaneously with advances in RNA sequencing, two factors have contributed to multimodal -omics: the development of analogous assays for chromatin accessibility and protein abundance, and the release of software infrastructure to quantify non-coding RNA molecules. If we run a scRNA-seq experiment, we get information about splicing for free. If we develop a slightly more sophisticated methodology, we can collect information about other modalities, either closer to gene regulation or to its effects.
Yet the correct way to jointly analyze such data is far from clear. Many data integration methods exist, but they tend to be descriptive and phenomenological, without encoding the causal relationships of the central dogma. For example, a single-nucleus RNA sequencing analysis will simply add spliced and unspliced RNA counts without accounting for their meaningful differences. One of the key goals of my Ph.D. work has been embracing multimodality: if we have interesting readouts, we should exploit them as much as possible.
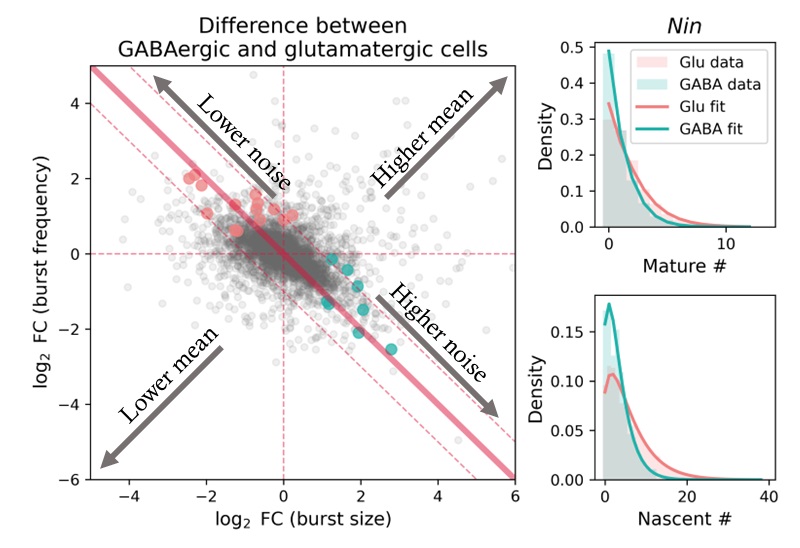
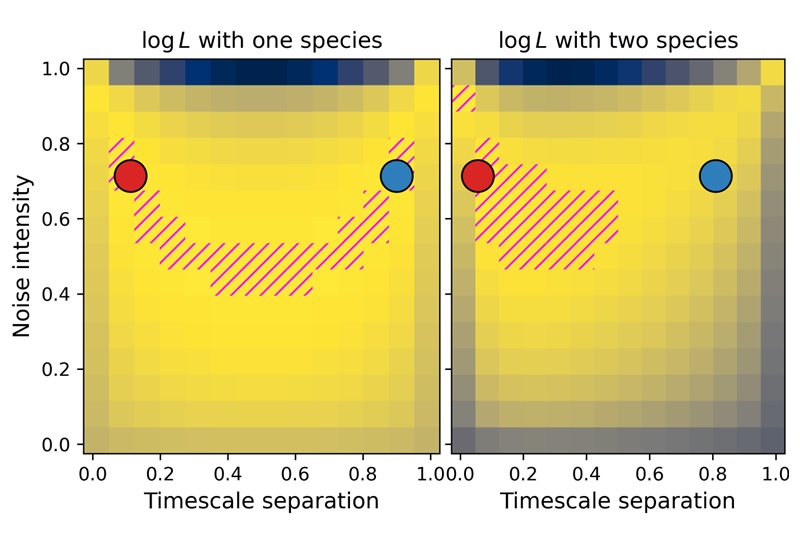
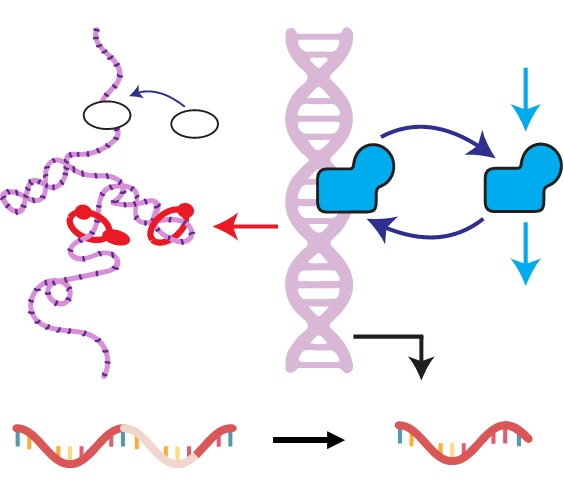
My theoretical work shows that whole is greater than the sum of its parts. For instance, having spliced and unspliced molecule counts allows us to distinguish between transcriptional mechanisms and regulatory strategies which would otherwise be indistinguishable using a single modality.
I am particularly interested in the interplay and connections between mechanistic and descriptive models. One of my recent projects, biVI, integrates spliced and unspliced data by endowing a machine learning framework with a biologically meaningful representation of the relationship between these molecules. In other words, the mechanistic worldview can naturally represent well-understood parts of the biophysics, whereas neural networks can represent currently uncharacterized “black box” parts.