Theory
Unifying sequencing analyses with stochastic theory
In brief: I develop, solve, and apply models that represent variability due to sequencing as well as biology, and use these models to understand the limitations of typical workflows.
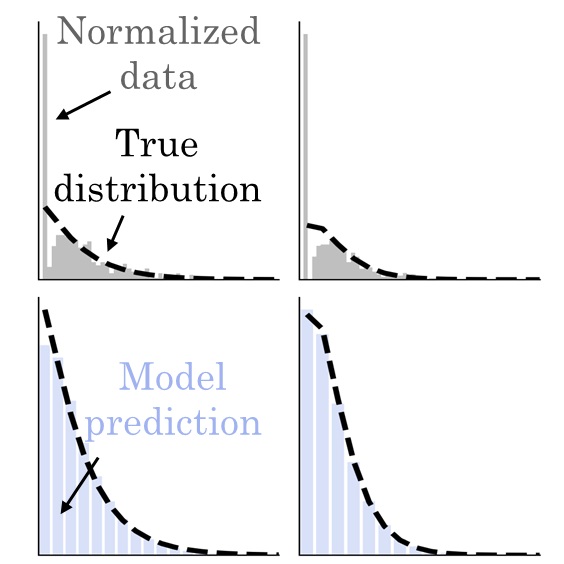
Single-cell workflows have been evolving at tremendous pace — yet data interpretation has been lagging behind in important ways. Conventional analyses take inspiration from the computer science field, using methods from signal processing and graph theory. But gene expression violates many of these methods’ assumptions: it is high-dimensional, discrete, and sparse. Even if the current methods work acceptably some of the time, I strive to characterize their limits and failure modes to understand how to build on them.
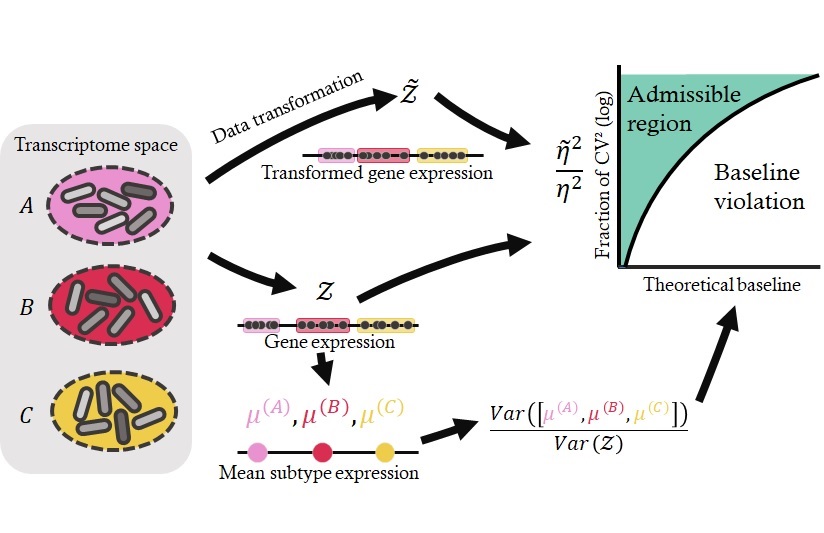
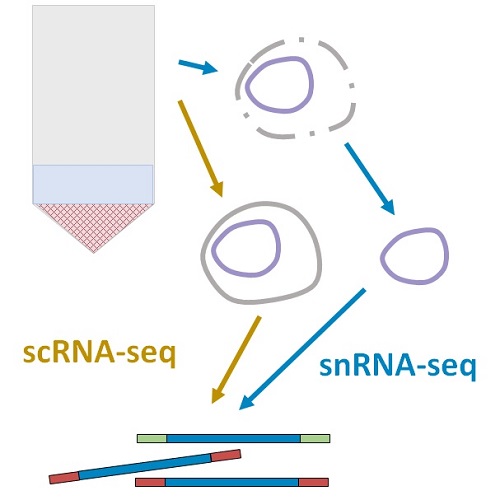
The observed experimental readouts combine biological variability due to cell type heterogeneity, transcriptional bursting, and single-molecule stochasticity, as well as technical variability due to imperfect sequencing, batch effects, and many as of yet uncharacterized sources of variation. To appropriately attribute and exploit differences in expression, we need to understand the contributions of biological and technical effects.
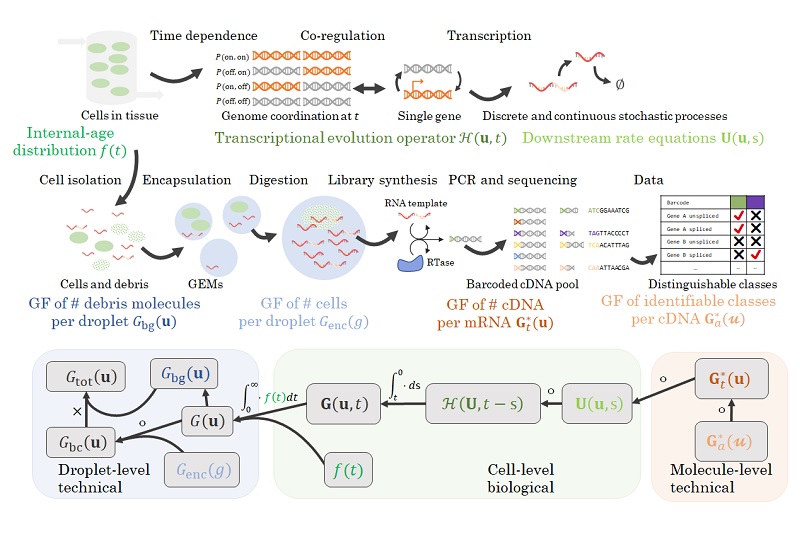
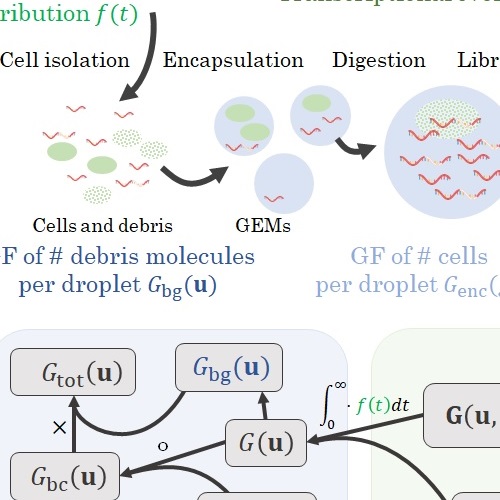
My Ph.D. work focused on developing a suite of interpretable, tractable models that combine the mechanisms of single-cell biology with the technology of single-cell RNA sequencing in a common stochastic framework. This approach brings the foundations of sequencing in line with theory developed for fluorescence transcriptomics, and confers two advantages.
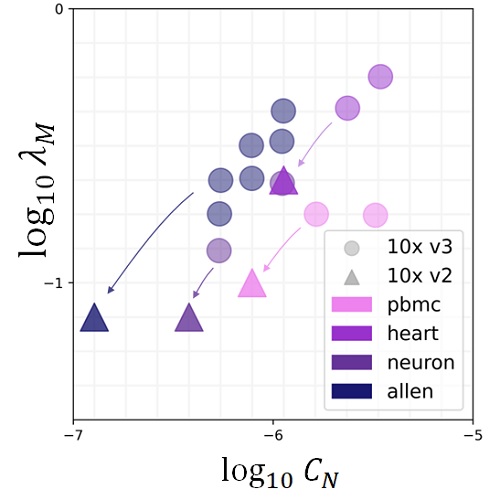
First, the mechanistic approach uses more of the data and provides more information than the descriptive approach. For example, instead of talking about differences in average gene expression, we can directly attribute those differences to changes in specific transcriptional parameters. To that end, I have implemented the Monod software package and used it to characterize subtle differences in biophysical processes which would otherwise be undetectable.
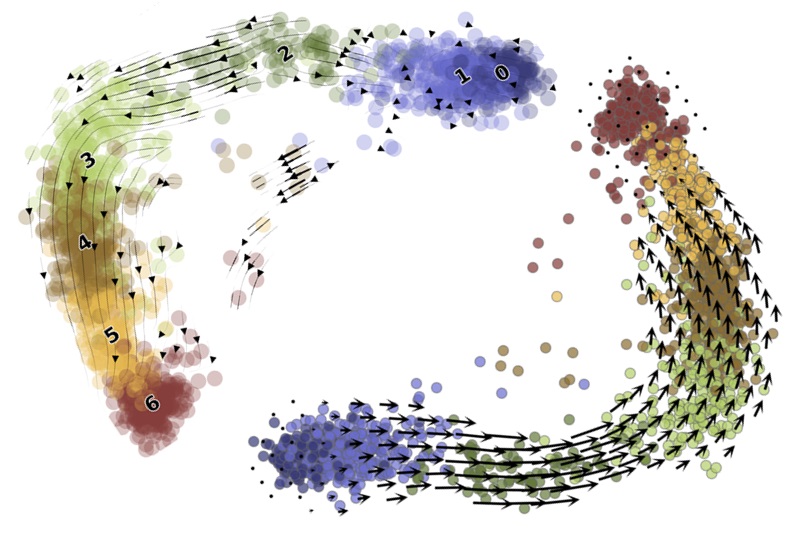
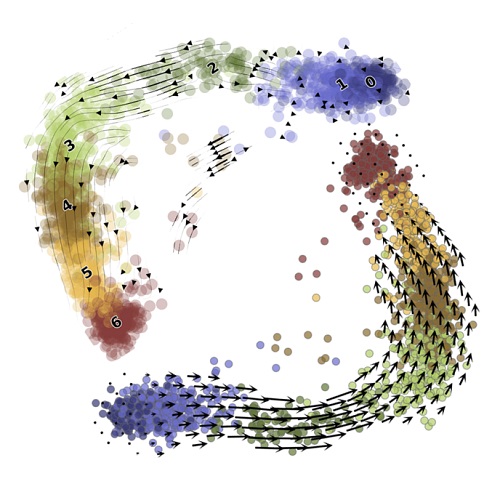
Second, it provides a principled way to ask questions of standard workflows, clarify their performance, and build better tools. I have used this approach to characterize the limitations of standard methods. For example, dimensionality reduction methods make severe assumptions, leading them to discard biologically meaningful variability. RNA velocity uses the outputs of dimensionality reduction workflows to visualize its results, leading to potentially flawed conclusions.